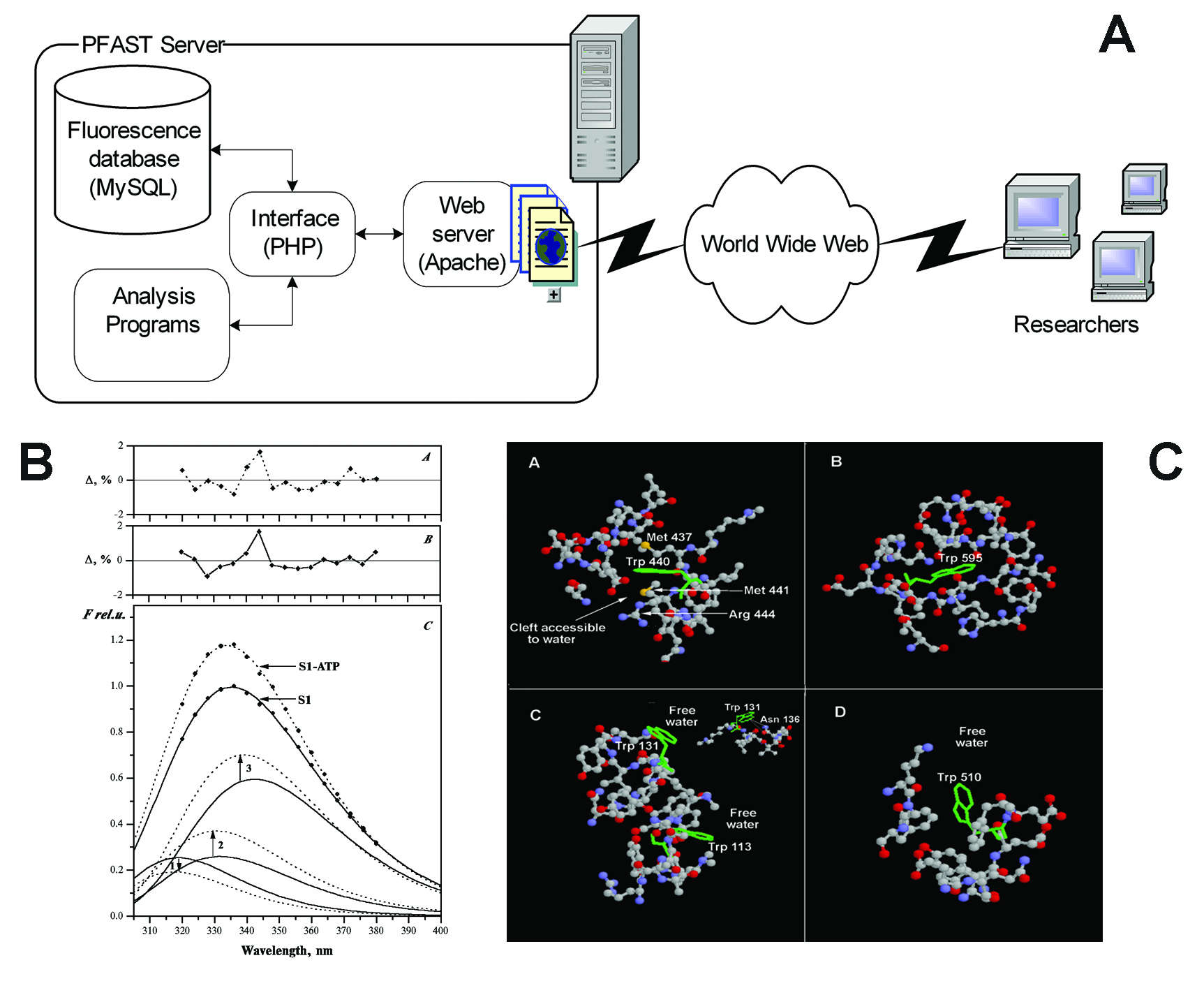
Protein Fluorescence
Our early work was directed toward establishing correlation between tryptophan fluorescence spectral properties and structural microenvironment of tryptophan residues in proteins. We have developed novel mathematical algorithms of spectral data analysis, which allow decomposing complex spectral profile of proteins (mostly containing more than one tryptophan residue) into individual components. Statistical approaches of multivariate analysis are employed to classify and assign spectral components to the protein structural features obtained in the result of analysis of atomic structures of proteins from Protein Data Bank. All algorithms are implemented into web-based toolkit PFAST: Protein Fluorescence And Structural Toolkit pfast.uri.edu